Quantum Insights: Dynamic Quantum Clustering (DQC) used for drug repurposing
(Original Document Here).
Summary
As part of RTTP's original 2018 p1RCC Hackthon, Quantum Insights' Marvin Weinstein and Bernard Chen ran their DQC program on both my DNA and TCGA’s DNA database of 20,000 matched samples across 33 cancer types. The goal of this "pan cancer analysis", to see if my cancer’s DNA was spatially adjacent to some other cancer types. If it was, the hope was that medications used in treating a “close” cancer could be used to treat my rare type of cancer. The results were inconclusive.
Fast forward to RTTP's and TRIcon's 2020 p1RCC hackathon. This time, UCLA’s Brian Shuch connected UCSF’s Max Meng and Tasha Lea with Yale’s Kaya Bilguvar. Tasha sent my blood and frozen tumor tissue to Kaya, who performed RNA-seq analysis which Mike D'Amour then post processed.. Next, Clemson's Colin Targonski, M. Reed Bender, Benjamin T. Shealy, Benafsh Husain, Melissa C. Smith and F. Alex Feltus ran this RNA-seq data through their TSPG program and found 18,000 genes that differentiated my tumor from normal kidney.
And this time, after running their DQC program on both Clemson's RNA gene list and TCGA’s RNA data, Marvin and Bernard discovered that my RNA data “clustered” close to TCGA’s Thyroid Cancer dataset. And unbeknowst to Quantum Insights, one of my siblings was diagnosed recently with both a lung carcinoid and thyroid cancer (The patient underwent an operation and is doing fine.).
Imagine that.
(Papillary) thyroid cancer and papillary kidney cancer links have been described previously (e.g. 1, 2). My hope is that there is biology behind this particular association. My next step is to enable discovery of a hard genetic link with a known oncogene or tumor suppressor.
An HTML version of Quantum Insights' Original Document is below. Thanks to all those who made the analysis possible. Anyone interested in helping explore the connection further please contact bill@rarekidneycancer.org.
Output of QuantumInsights.io's clustering algorithm
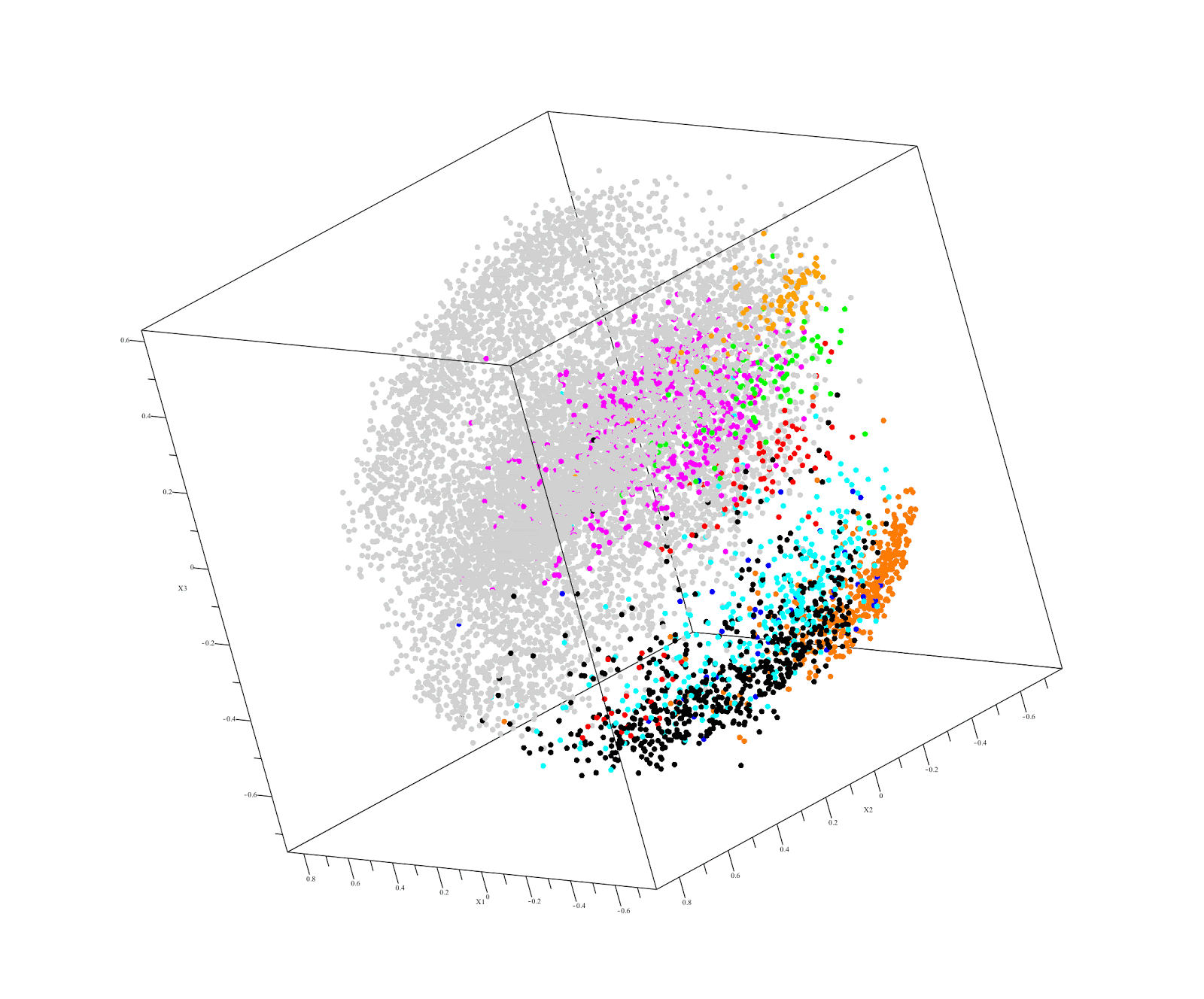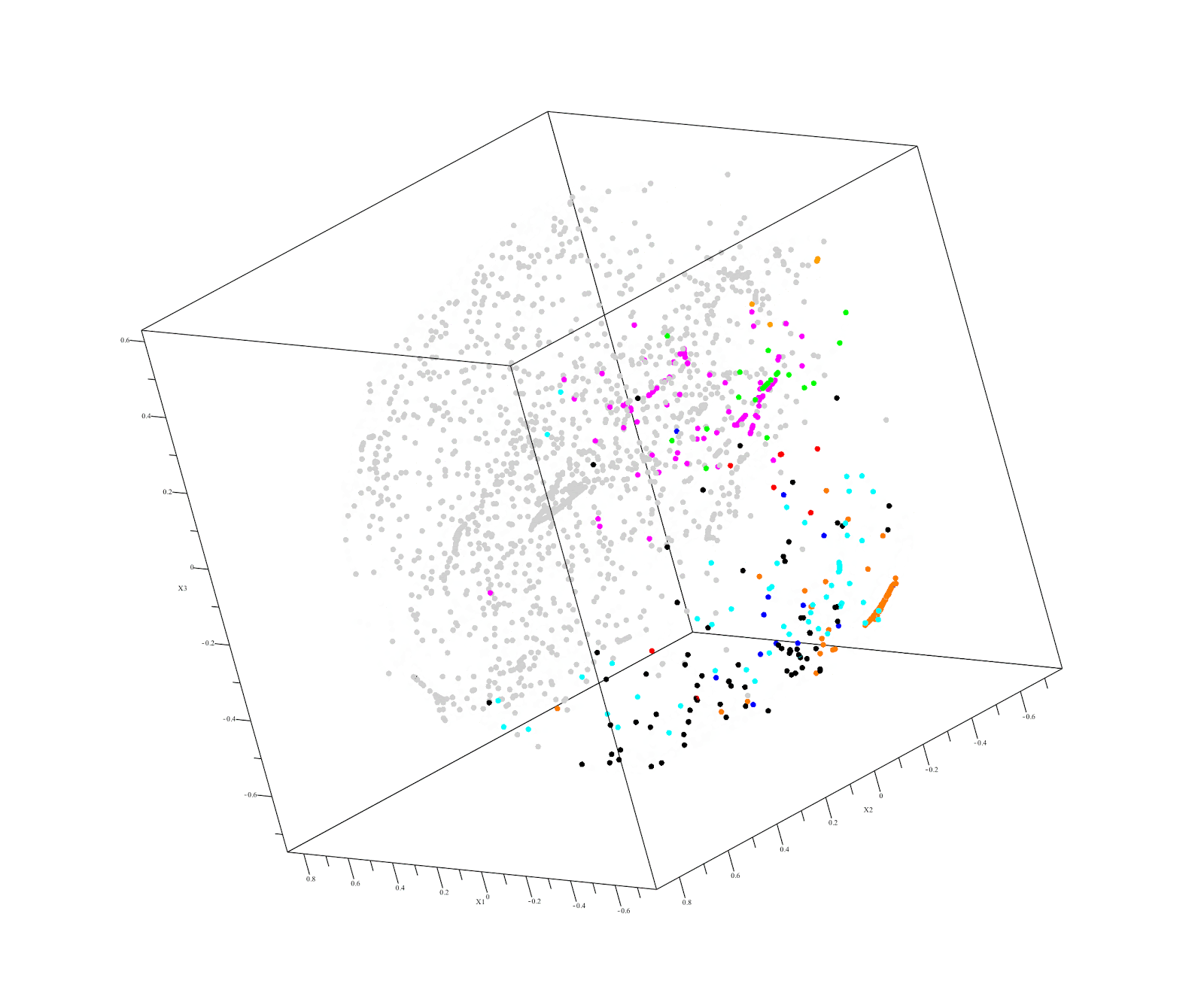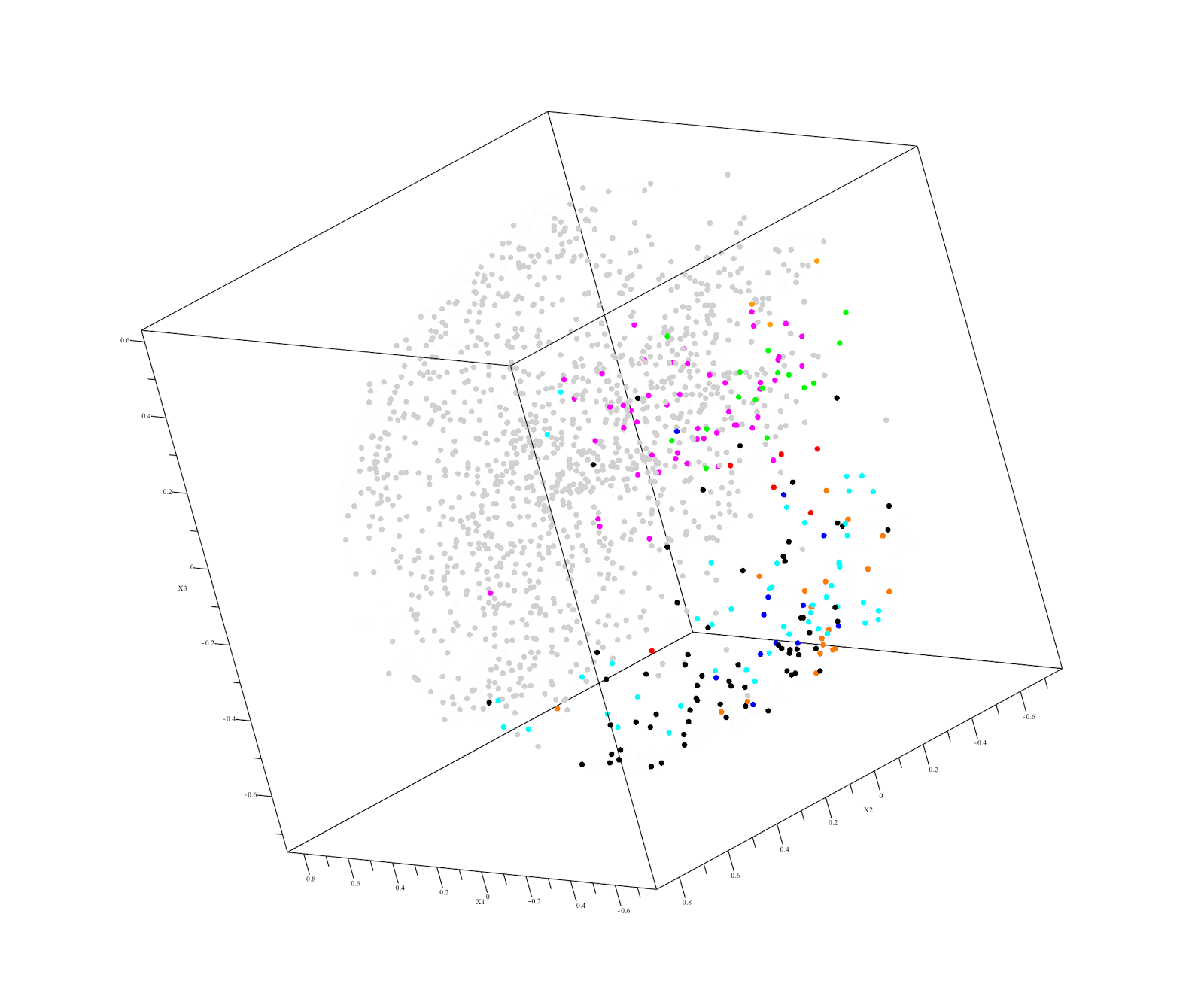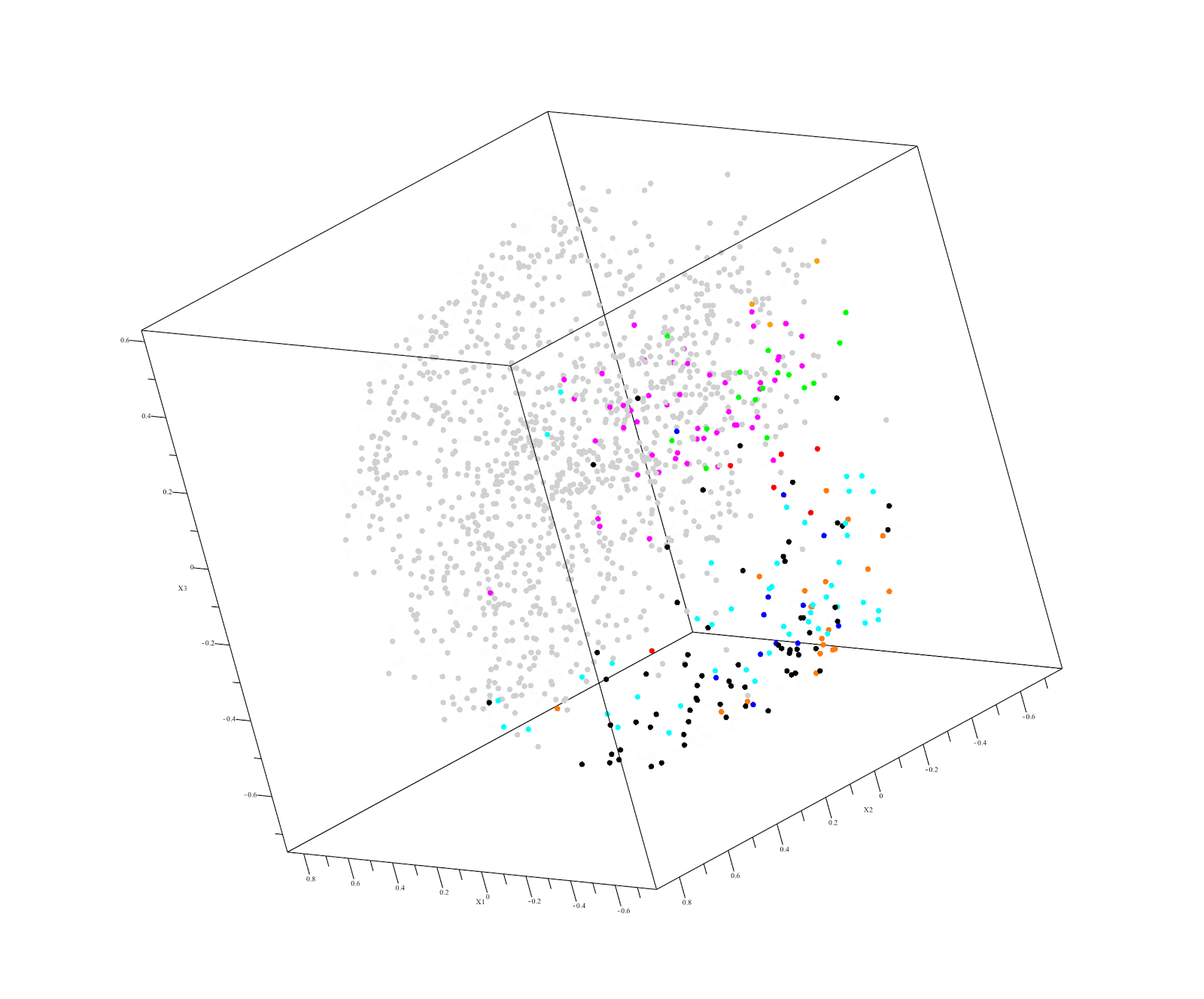

Executive Summary:
A potential, less costly, alternative to the classical drug discovery pipeline is drug repurposing. Repurposing - the process of identifying an already approved drug that can work on a disease other than that the disease it was designed to attack – can increase the market for an existing drug. But, from a human point of view, there is another virtue of this approach. It can save lives by identifying drug candidates for treating rare or orphan diseases. Namely, diseases that – for economic reasons – do not get much attention from large pharmaceutical companies.
Quantum Insights (QI) has developed a method that uses RNAseq data from TCGA (and other possible sources) to identify drugs most likely to be successfully repurposed to treat new diseases. The method leverages DQC’s ability to identify diagnostic signatures - using a density mapping approach driven by quantum mechanics principles (The technical paper on the method is available here) - to create a similarity distance between the diseases for which TCGA RNAseq data is available. We then focus on a specific drug – or disease - in question and use the similarity distance map to find the diseases that are closest to this disease. Finally, we determine which genes play the most important role in defining the similarity distance and examine whether there is evidence if any of these genes are known targets of the drug to be repurposed. If we find a match between a drug and a disease candidate for which that drug can be repurposed, then we look for clinical evidence that our prediction is correct. Clearly the same approach can be used to find candidate drugs to treat a disease for which there is no known current treatment.
A Case Study: KIRP – an orphan kidney cancer
Kidney renal papillary cell carcinoma (KIRP or PRCC) is a renal cell cancer with poor prognosis under current treatments. It comprises about 10-15% of all kidney cancers. At present there are not good treatments for KIRP. Our challenge was to find an existing drug that is likely to help treat a patient diagnosed with KIRP using the RNAseq expression data for the patient’s tumor. We began by analyzing RNAseq data – obtained from TCGA (40 cancers, n=10,000, 60,000 genes) - and restricted the genes to the 18,000 genes present in a patient’s poor quality KIRP sample. Our similarity distance analysis identified several non-kidney cancers closest to KIRP. These were:
- Liver hepatocellular carcinoma (LIHC)
- Thyroid carcinoma (THCA)
- Cholangiocarcinoma (CHOL)
- Uveal melanoma (UVM)
- Adrenocortical carcinoma (ACC)
We next identified existing, FDA-approved, drugs to treat each disease.
- CHOL, UVM and ACC did not have FDA-approved drugs and were excluded from the rest of the analysis.
- LIHC and THCA have, in total, 10 FDA-approved drugs. (Listed in Appendix A)
Next, using the Open Targets website (https://www.targetvalidation.org), we identified the target for each approved drug, resulting in 47 target genes (listed in Appendix B).
- Three genes (Programmed Cell Death 1; PDCD1, Immunoglobulin Heavy Constant Epsilon; IGHE, and Epherin Receptor B6; EPHB6) were not included in the TCGA list of 18,000 genes and were excluded from the rest of the analysis.
- We calculated the gene expression difference for KIRP vs “healthy kidney” samples as defined in the TCGA.
- There are 30 genes where the difference was significant, but only 4 genes that were overexpressed in KIRP relative to healthy kidney cells: Proto-Oncogene Receptor Tyrosine Kinase(MET), Transmembrane Protein 4 (TMEM43 – aka Luma ), Sarcoma gene (SRC), Erb-B2 Receptor Tyrosine Kinase (ERBB3 ). We focused on overexpressed genes because the action of these four drugs is to inhibit their targets.
Presently, we have no clinical data that tells us if these drugs will work on KIRP. However, it turns out that for the drugs that target MET, SRC, TMEM43, and ERBB3 (Cabozantinib, Nivolumab, and Vandetanib) there are several clinical trials testing the hypothesis that these drugs work on KIRP2:
-
3 trials including Cabozantinib: NCT02761057, NCT03685448, NCT03635892
-
2 trials including Nivolumab: NCT03177239, NCT03635892 (this is a duplicate trial: Cabozantinib + Nivolumab)
-
1 trial including Vandetanib: NCT02495103
When these trials complete we will be able to assess the validity of our prediction.
2 Searching Clinicaltrials.gov on “Kidney renal papillary cell carcinoma” (must target papillary vs other kidney cancers
Appendix A: Drugs approved for LIHC and THCA
| LIHC | liver hepatocellular carcinoma | Target Engagement |
| Cabometyx, Cometriq | Cabozantinib | MET, KDR, MAD2L1BP |
| Cyramza | Ramucirumab | KDR |
| Keytruda | Pembrolizumab | PDCD1, IGHE |
| Lenvima | Lenvatinib Mesylate | FLT4, KDR, FLT1 |
| Nexavar | Sorafenib Tosylate | PDGFRB, RAF1, KIT, RET, BRAF, FLT1, KDR, FLT3, FLT4 |
| Opdivo | Nivolumab | PDCD1, TMEM43 |
| Stivarga | Regorafenib | ABL1, RAF1, KIT, BRAF, DDR2, FGFR2, PDGFRA, PDGFRB, FGFR1, RET, KDR, FLT4, TEK, FLT1, FRK, MAPK11, EPHA2, ABCB1, CYP3A4 |
| THCA | Thyroid Cancer | |
| Cabometyx, Cometriq | Cabozantinib | MET, KDR, MAD2L1BP |
| Caprelsa | Vandetanib | ERBB3, EPHB2, SRC, EGFR, RET, EPHA3, ERBB4, EPHB4, ERBB2, EPHA7, TEK, KDR, EPHA2, PTK6, EPHA4, EPHB1, FLT4, EPHB3, EPHA10, EPHB6, FLT1, EPHA8, EPHA6, EPHA5, VEGFA, CYP3A4 |
| Tafinlar | Dabrafenib Mesylate | BRAF, CPB2 |
| Lenvima | Lenvatinib Mesylate | FLT4, KRD, FLT1 |
| Mekinist | Trametinib | MAP2K1, MAP2K2, CYP3A4 |
| Nexavar | Sorafenib Tosylate | PDGFRB, RAF1, KIT, RET, BRAF, FLT1, KDR, FLT3, FLT4 |
The 10 drugs approved for LIHC and THCA are:
- Cabozantinib
- Ramucirumab
- Pembrolizumab
- Lenvatinib Mesylate
- Sorafenib Tosylate
- Nivolumab
- Regorafenib
- Vandetanib
- Dabrafenib Mesylate
- Trametinib
Appendix B: 30 Genes differentiating KIRP from Healthy Kidney
CYP3A4 FLT4 DDR2 FRK EPHA3 KDR EPHA4 KIT EPHA5 MAP2K1 EPHA6 MAPK11 EPHA8 MET EPHB1 PDGFRA EPHB3 PDGFRB EPHB4 RAF1 ERBB2 RET ERBB3 SRC ERBB4 TEK FGFR2 TMEM43 FLT1 VEGFA
Note: Genes in bold are overexpressed relative to healthy kidney cells (all were p<0.00136). The other genes are significantly under expressed.
Appendix C: Open Questions
● Out of 30 genes that are significantly different between KIRP our tumor adjacent “healthy” kidney cells, only 4 were over-expressed in KIRP. Many other genes were significantly suppressed, is a 1:6.5 ratio of over-to-under expression surprising?
○ Is this small number because tumor adjacent cells are not really healthy cells? Quantum Insights is looking for “true healthy” cells in order to re-run this analysis.
● Are there other ways (besides KIRP/healthy kidney ratio) to determine if a subset of the 10 drugs will have a positive effect on KIRP?


Add new comment