Introduction
My name is Bill Paseman. I have an incurable, terminal (but fortunately indolent) Kidney Cancer called "papillary type 1 RCC" (as well as a brain cancer). In 2017, rare kidney cancer researcher Dr. Laurence Albiges noted that OS (Overall Survival) for Papillary RCC had not increased in more than a decade (slide 5). So in 2018, I started looking into new ways of doing cancer research.
As I have mentioned elsewhere, being a terminal cancer patient gives me a sense of urgency not typically present in an ordinary researcher. As such, I am not particularly interested in the learning the tools used by cancer researchers since they likely can use them better than I. Nor am I interested in developing tools for cancer researchers to use, since there are also better tool developers than I out there. Instead, in the words of Douglas Engelbart, I’m interested in “Improving how Cancer Researchers Improve”. This requires me to get my arms around the whole cancer research process.
In that regard, I was inspired in 2018 by the work of NIH’s Ben Busby and SV.ai’s Pete Kane who run “hackathons": weekend events attracting young researchers who concentrate on a single topic in parallel over three days and deliver a report at the end. Researchers typically form teams including Biologists, Bioinformaticians and Computer Scientists. A complete research process in miniature. Hackathons discussed here are “Patient Centered”, meaning Patients themselves provide their data for study (2018 and 2020) as well as host and maintain control of the event.
The focus of my effort is to make hackathons "self improving processes" by:
- Incorporating “Game Elements” such as teams, levels, scores and leaderboards.
- Treating research teams as AI style "computational objects", which lets me apply AI techniques such as reinforcement learning and Ensemble learning to improve the research outcome.
Here, I summarize the results of these efforts.
2018 p1RCC Hackathon Teams

Shown above are the seventeen teams that accepted Pete and my invitations to the 2018 San Francisco p1RCC hackathon. They came from Stanford, Harvard, UCSF, Rutgers, Clemson (which has been a great supporter) and many other institutions.
2018 Hackathon Game Flow and Scoring
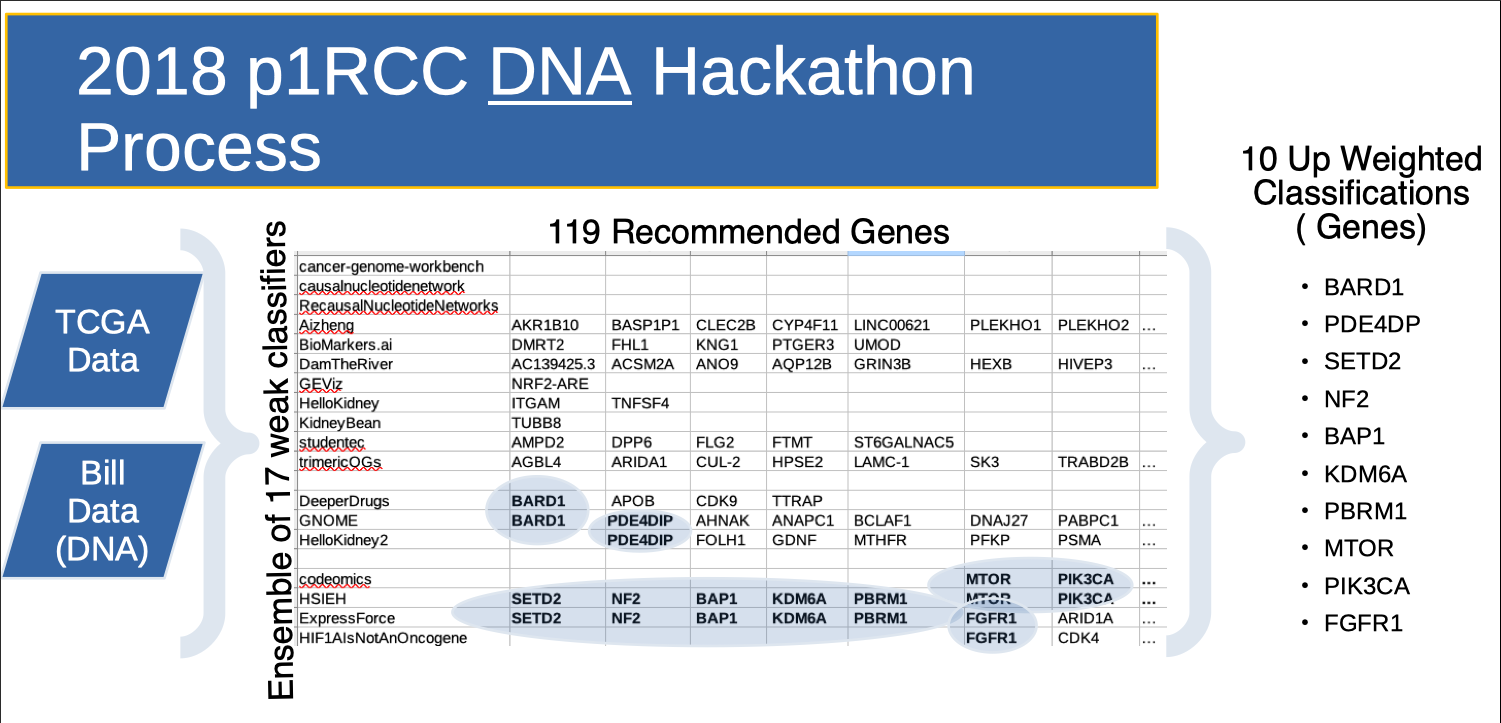
The slide above illustrates the 2018 hackathon game flow. An ensemble of teams was given WGS (Whole Genome Sequencing) of my normal blood and my FFPE tumor tissue and access to TCGA data. During the initial diagnostic "game level", each team researched the data in parallel to identify "Genes of Interest" (GOI). To aid in prioritizing effort during the therapeutic "game level", each GOI was "scored" two ways. The first score used “known results”. That is, if the team discovered a gene on a cancer researcher's (Dr. James Hsieh's) curation list, they scored! (But not particularly highly since these genes were already known in the papillary kidney cancer literature, and teams could find them via Google.) This produced genes like NF2, MTOR and BAP1. The second score used “results overlap”. That is, if two teams working independently each recommend the same gene, they scored! The thought here is that if two paths of research lead to the same gene, it ought be investigated. This produced genes like PDE4DIP, BARD1 and FGFR1. Typically, teams produced therapeutic recommendations based on the genes they themselves identified. As such, the diagnostic "scores" served to add weight to the therapeutic recommendations.
In addition to "Genes of Interest", this hackathon produced a paper written by Clemson's William Poehlman in conjunction with Dr. James Hsieh.
2020 Hackathon Game Flow
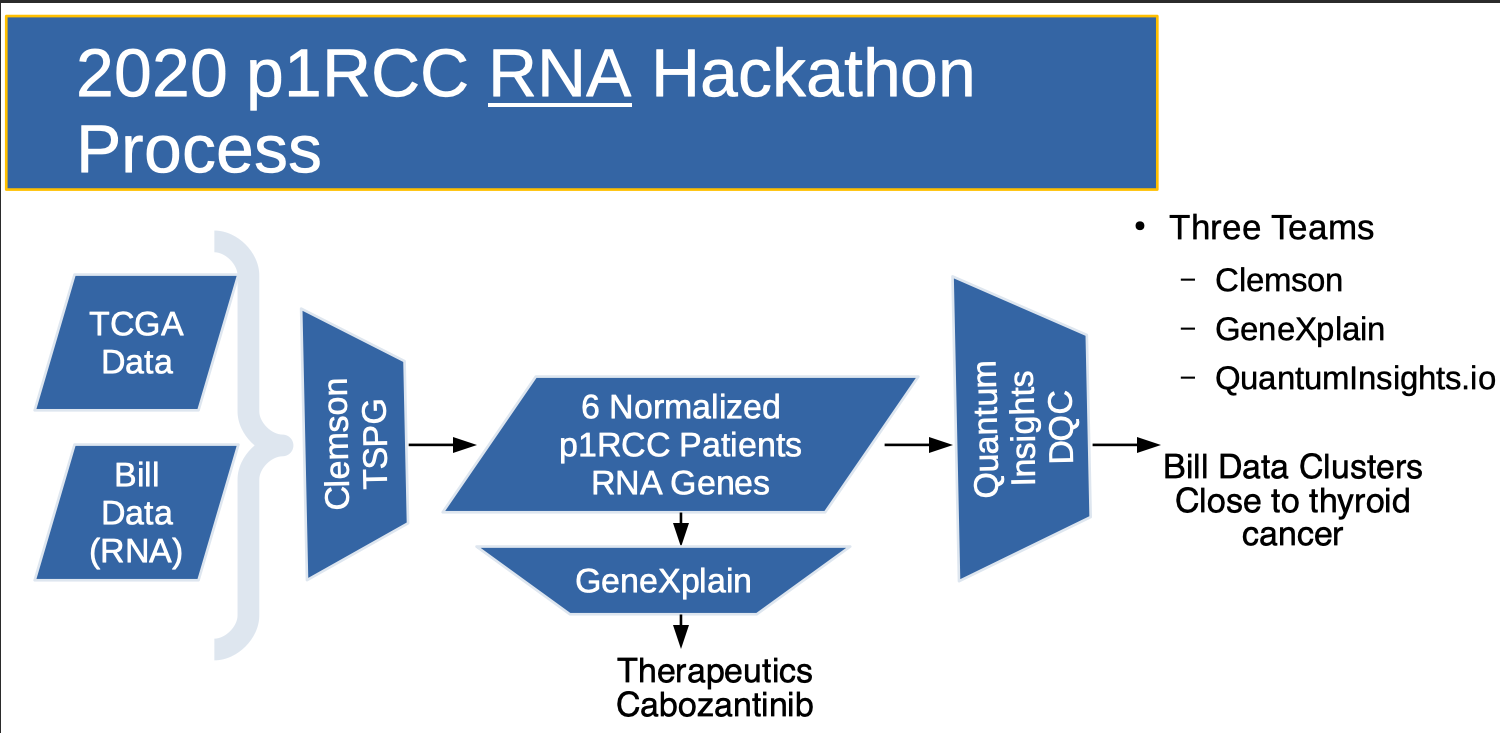
In the 2020 hackathon, at the beginning of the Covid-19 pandemic, we had light turnout. The 2020 Flow (above) was similar to 2018, but results were more open ended. Great foundational work was done by Clemson's Reed Bender who created a “perturbation list” that registered the expression level "shift" between my tumor and my "kidney normal" RNA-seq data. This perturbation list is sorted. High positive values indicate that the gene is over expressed in the tumor. High negative values indicate that the gene is under expressed in the tumor. This enabled a therapeutic recommendation (Cabozantinib) by the GeneXplain team.
Using Siblings and Parents as Controls
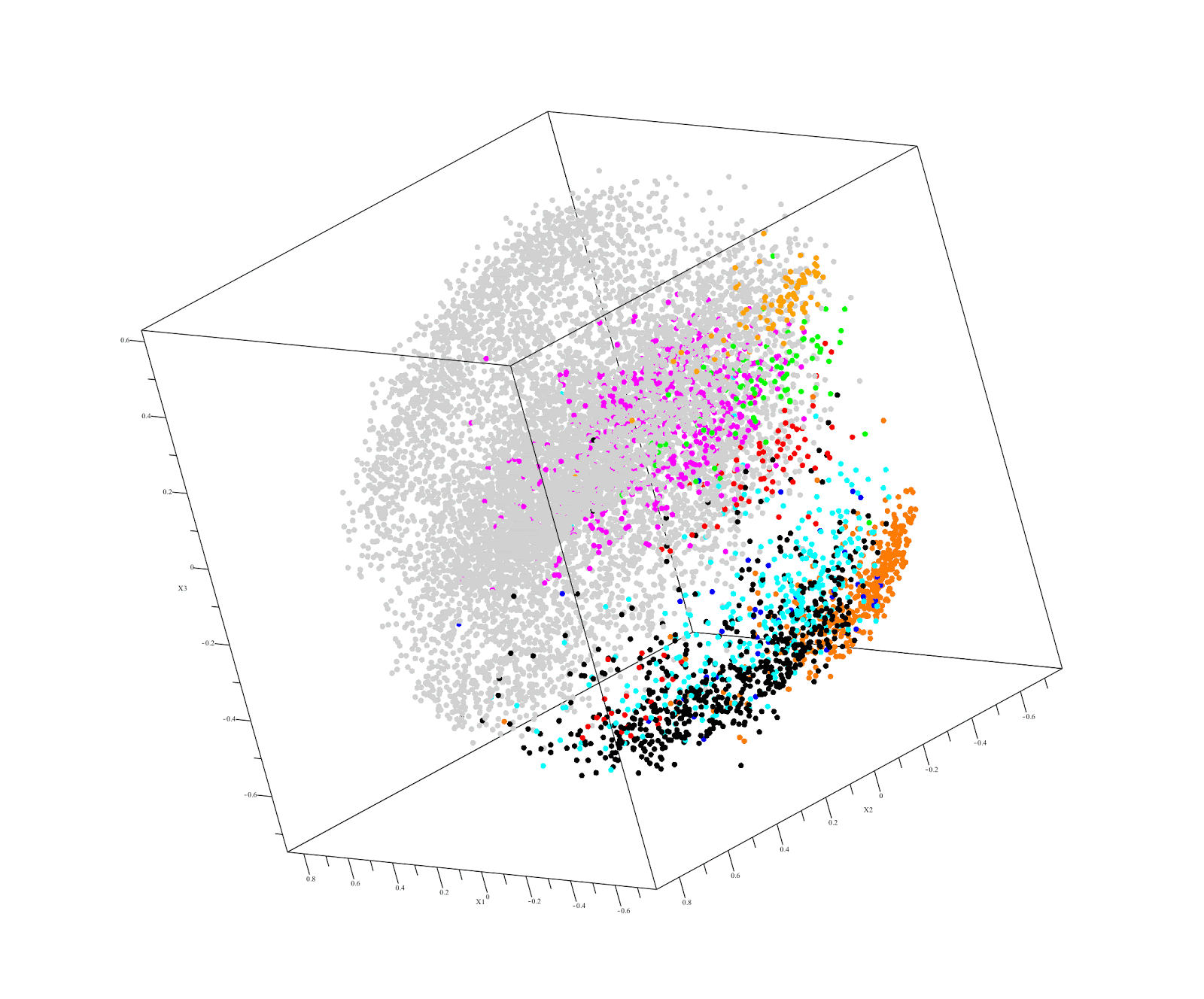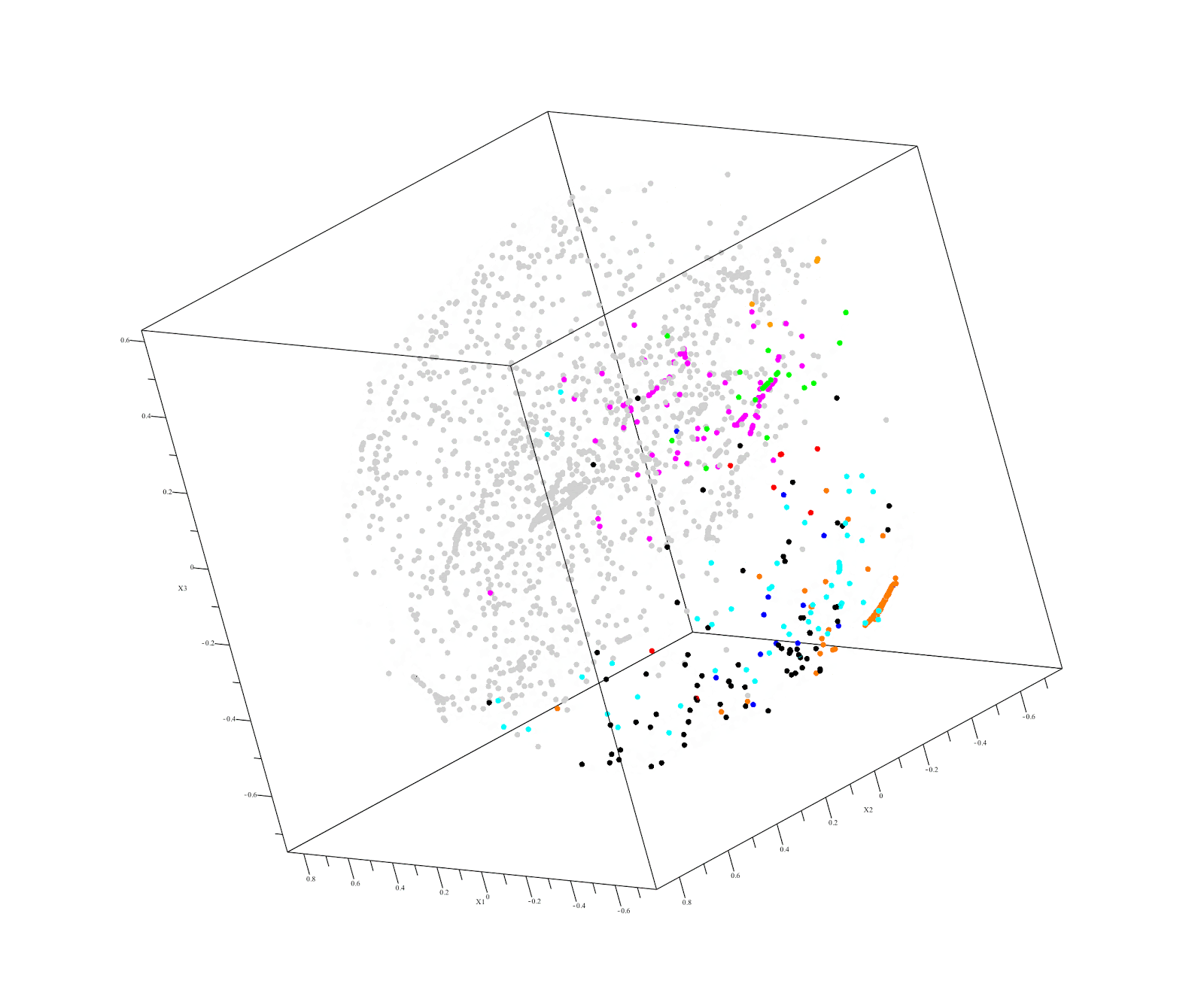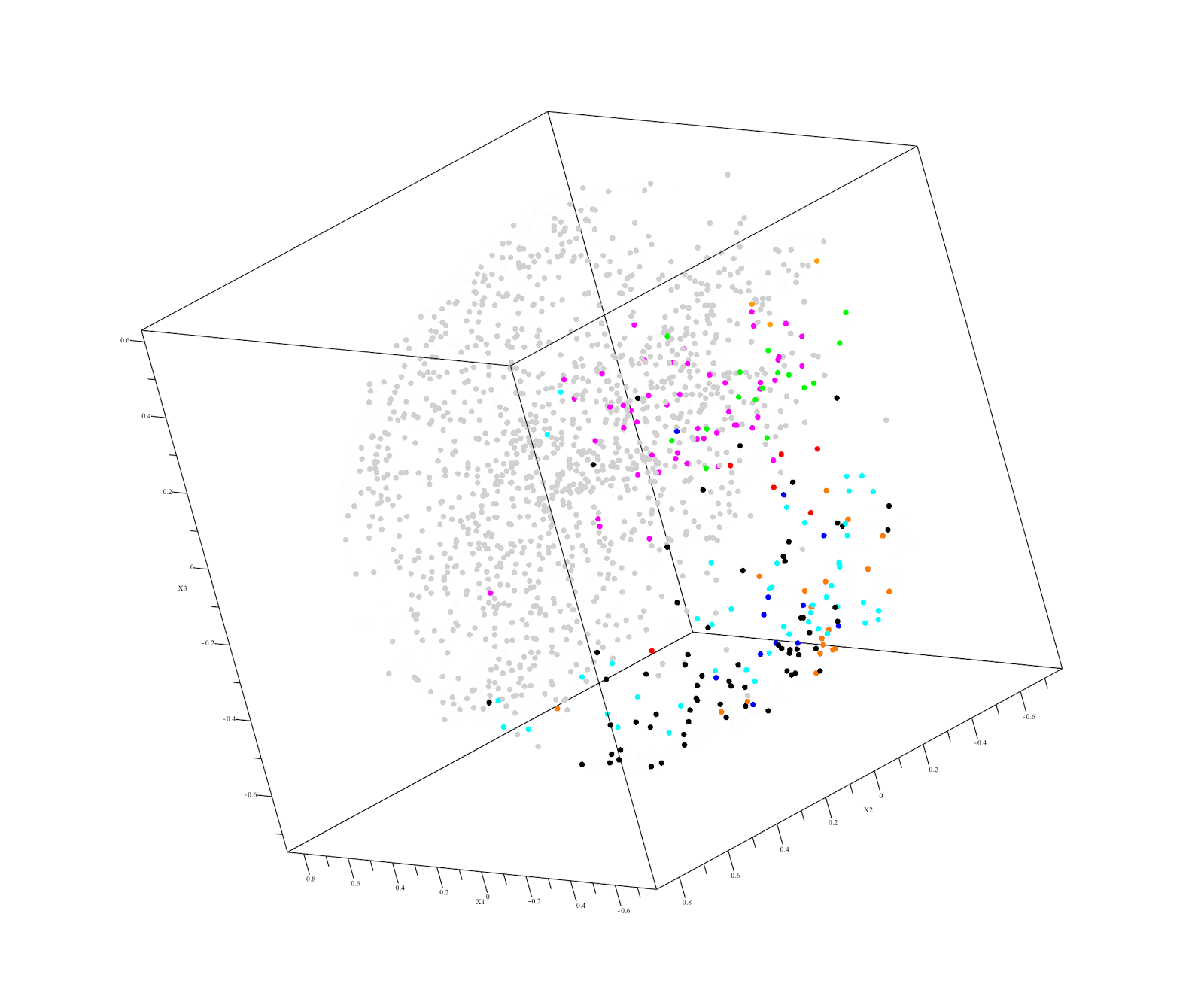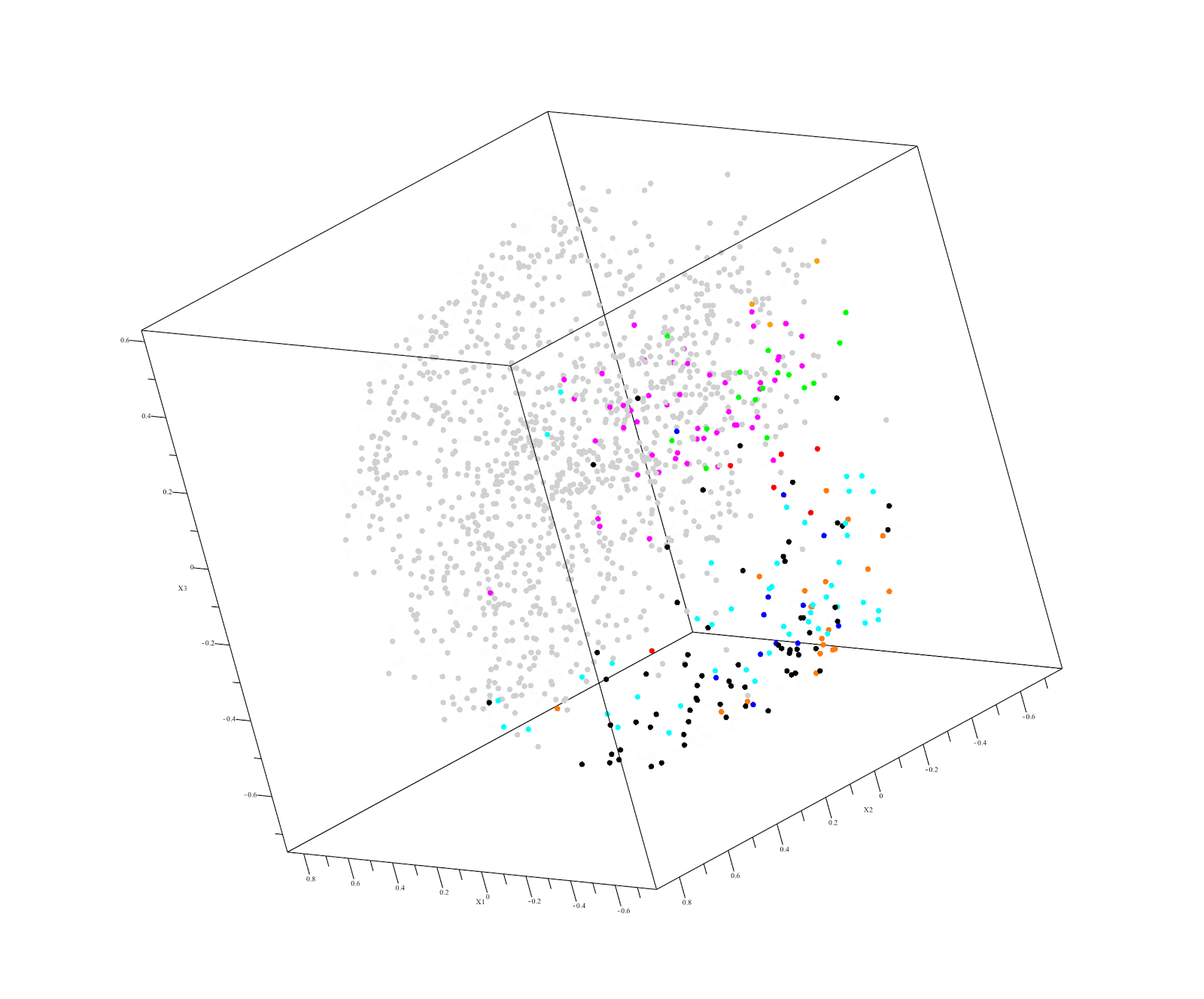

This raises the possibility of using familial DNA as a "control" in our next hackathon. This approach is already used by Derya Karaarsian (here and here). Also, in a recent Desmoid tumor hackathon, Vanessa discovered that her father had keloids, which pushed her research in a whole new direction.
2020 Scoring of 2018 Results
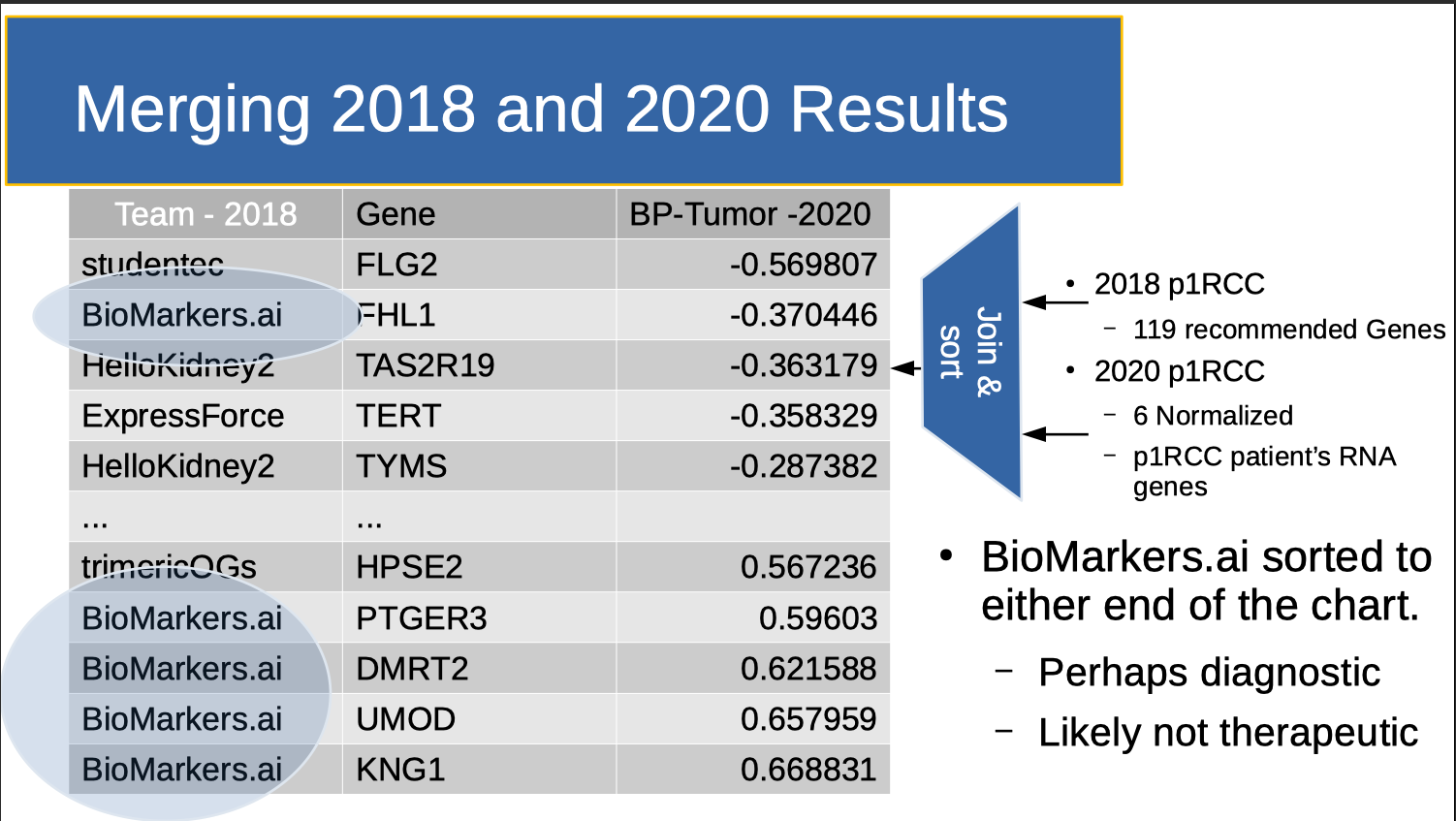
Note that Reed’s perturbation data was not available in 2018. So post hoc, I scored the 2018 data a third way, asking where the “genes of interest” fell on the perturbation list (above Figure). Note that applying this scoring approach post hoc essentially meant that perturbation functioned as a “holdout set”.
Of the seventeen 2018 teams, Rutger’s Saed Sayad knocked this third score “out of the park”. In particular, the genes he recommended investigating included:
- KNG1 - which is related to blood coagulation, and may explain a DVT I had while therapeutic on warfarin.
- UMOD - which is the focus of the CKD community.
- FHL1- which is an indicator for petrochemical exposure, and may have occurred as a result of my prior jobs in refineries and drilling rigs.
Using Differential Expression to create a LeaderBoard
| Team - 2018 | Gene | BP-Tumor -2020 | Approach |
| studentec | FLG2 | -0.569807 | https://github.com/SVAI/studentec |
| BioMarkers.ai | FHL1 | -0.370446 | https://github.com/SVAI/Biomarkers.AI |
| HelloKidney2 | TAS2R19 | -0.363179 | https://github.com/SVAI/HelloKidney2 |
| ExpressForce | TERT | -0.358329 | https://github.com/SVAI/ExpressForce |
| HelloKidney2 | TYMS | -0.287382 | https://github.com/SVAI/HelloKidney2 |
| ... | ... | ||
| trimericOGs | HPSE2 | 0.567236 | https://github.com/SVAI/trimericOGs |
| BioMarkers.ai | PTGER3 | 0.59603 | https://github.com/SVAI/Biomarkers.AI |
| BioMarkers.ai | DMRT2 | 0.621588 | https://github.com/SVAI/Biomarkers.AI |
| BioMarkers.ai | UMOD | 0.657959 | https://github.com/SVAI/Biomarkers.AI |
| BioMarkers.ai | KNG1 | 0.668831 | https://github.com/SVAI/Biomarkers.AI |
Note that Reed's perturbation data can can be used to create a "LeaderBoard" that shares all research approaches for all genes with all participants. For example, here is the Biomarkers.ai Leaderboard Entry from the above table. How did Biomarkers get such good results? Basically, they used my DNA (which has a weak statistical signal) to look up RNA (which has a strong statistical signal) in GEO. This strong signal allowed them to use a relatively simple technique (Linear Discriminant Analysis) to separate out "Genes of Interest". Using a gamified format to share "best of breed" research results is educational, novel, important and critically, fun.
Therapeutic Options
Dr. Sayad then used his Bioada tools to suggest therapeutic options, including Valproic acid and Baicalein.
Ensemble Learning
In AI, Ensemble learning is the process by which multiple models, such as classifiers or experts, are strategically generated and combined to solve a particular computational intelligence problem. Ensemble learning is primarily used to improve the (classification, prediction, function approximation, etc.) performace of the group. Ensemble analysis showed that 2018 teams using research (RNA) versus clinical(DNA) data produced better results, that smaller teams had better results (See also "Can Science be too Big?") and that smaller teams took more risk. This relationship between risk and return is also present in portfolios of financial assets as well. We discuss this more here.
Future Work
|
|
SV.ai |
TRIcon |
Clemson |
Stanford |
|
|
2018 |
2020 |
2022 |
202x |
|
Bill |
x |
x |
x |
x |
|
Gigi |
|
|
x |
x |
|
Patient X |
|
|
|
x |
Clemson is planning a hackathon in March of 2022 for Gigi, who suffers from hypophosphatasia. One research track will also include a group working on my p1RCC data. We hope to hold a hackathon at Stanford after that. Notice the opportunity we have here for network effects. The opportunity to bring together researchers of rare diseases with their patients. The opportunity to create a social network for social good.
Summary
I investigated “Patient Centered” research approaches that incorporate “Game Elements” and “Ensemble Learning”.
“Patient Centered” means that:
- Patients view themselves as having a “rare disease” that is not served well by cohort analysis. We hope to use sibling and parent genetic data as a "control" in future events.
- Patients themselves host and maintain control of the event and are responsible for providing their own data.
- Data Control allows patients to create a current, longitudinal record over time for each subsequent hackathon as their disease develops.
“Game Elements” means that:
- Hackathon participants are divided up into teams.
- The Game has “levels” which include diagnosis and therapeutic recommendations.
- Team’s results are “scored” which helps the Patient prioritize future research approaches.
- Scores can be posted on a LeaderBoard, which allows sharing of Research Approaches.
Treating Research Teams as formal computational objects lets us apply an “Ensemble Learning” technique called "bucket of models".
- For each model m in the bucket:
- Do c times: (where 'c' is some constant)
- Randomly divide the training dataset into two datasets: A, and B.
- Train m with A
- Test m with B
- Select the model that obtains the highest average score
We are considering other ensemble techniques, and novel ways to do clinical trials.
If you want to learn further about this work, please contact bill@rarekidneycancer.org
This is a simplified version of an earlier post.


Add new comment